This page was generated from
jupyter-notebook/nb_04batch-inversion.ipynb.
Interactive online version:
 .
Download notebook.
.
Download notebook.
Batch inversion
Batch inversion is the fact to allow to invert multiple surveys in once. Here we reuse a lot of stuff from the time-lapse implementation.
[1]:
%matplotlib inline
import warnings
warnings.filterwarnings('ignore')
import os
import sys
import matplotlib.pyplot as plt
sys.path.append((os.path.relpath('../src/'))) # add here the relative path of the API folder
testdir = '../src/examples/dc-2d-timelapse/'
from resipy import Project
API path = /media/jkl/data/phd/resipy/src/resipy
ResIPy version = 3.4.6
cR2.exe found and up to date.
R3t.exe found and up to date.
cR3t.exe found and up to date.
[2]:
k = Project(typ='R2')
k.createBatchSurvey(testdir + 'data/')
Working directory is: /media/jkl/data/phd/resipy/src/resipy
clearing dirname
3/3 imported
[3]:
fig, ax = plt.subplots()
k.fitErrorPwl(ax=ax)
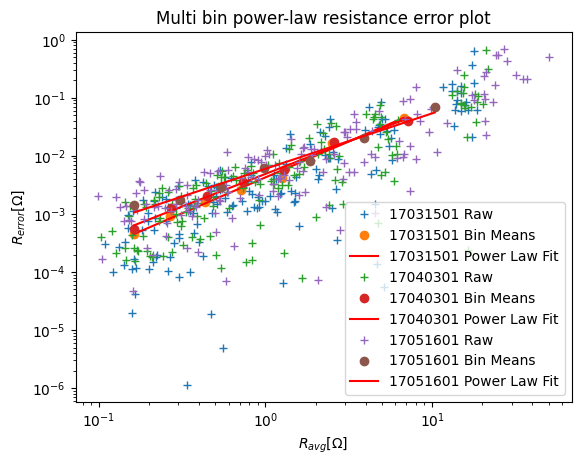
And inversion as usual.
[4]:
k.invert(parallel=True) # as for time-lapse, we can run the inversion in parallel
# (this will makes the live outputs a bit funny but should be faster overall.)
Creating triangular mesh...done (1786 elements)
Writing .in file and protocol.dat... done
--------------------- MAIN INVERSION ------------------
0/3 inversions completedors
>> R 2 R e s i s t i v i t y I n v e r s i o n v4.10 <<
>> D a t e : 03 - 12 - 2023
>> My beautiful survey
>> I n v e r s e S o l u t i o n S e l e c t e d <<
>> Determining storage needed for finite element conductance matrix
>> R 2 R e s i s t i v i t y I n v e r s i o n v4.10 <<
>> D a t e : 03 - 12 - 2023
>> My beautiful survey
>> I n v e r s e S o l u t i o n S e l e c t e d <<
>> R 2 R e s i s t i v i t y I n v e r s i o n v4.10 <<
>> D a t e : 03 - 12 - 2023
>> My beautiful survey
>> I n v e r s e S o l u t i o n S e l e c t e d <<
>> Generating index array for finite element conductance matrix
>> Determining storage needed for finite element conductance matrix
>> Determining storage needed for finite element conductance matrix
>> Reading start resistivity from res0.dat
>> Generating index array for finite element conductance matrix
>> Generating index array for finite element conductance matrix
>> R e g u l a r i s e d T y p e <<
>> L i n e a r F i l t e r <<
>> L o g - D a t a I n v e r s i o n <<
>> N o r m a l R e g u l a r i s a t i o n <<
>> D a t a w e i g h t s w i l l b e m o d i f i e d <<
>> Reading start resistivity from res0.dat
>> Reading start resistivity from res0.dat
>> R e g u l a r i s e d T y p e <<
>> L i n e a r F i l t e r <<
>> L o g - D a t a I n v e r s i o n <<
>> N o r m a l R e g u l a r i s a t i o n <<
>> D a t a w e i g h t s w i l l b e m o d i f i e d <<
>> R e g u l a r i s e d T y p e <<
>> L i n e a r F i l t e r <<
>> L o g - D a t a I n v e r s i o n <<
>> N o r m a l R e g u l a r i s a t i o n <<
>> D a t a w e i g h t s w i l l b e m o d i f i e d <<
Processing dataset 1
Measurements read: 190 Measurements rejected: 0
Geometric mean of apparent resistivities: 0.51162E+02
>> Total Memory required is: 0.003 Gb
Iteration 1
Processing dataset 1
Measurements read: 190 Measurements rejected: 0
Geometric mean of apparent resistivities: 0.52987E+02
>> Total Memory required is: 0.003 Gb
Processing dataset 1
Measurements read: 190 Measurements rejected: 0
Geometric mean of apparent resistivities: 0.70466E+02
>> Total Memory required is: 0.003 Gb
Iteration 1
Iteration 1
Initial RMS Misfit: 30.43 Number of data ignored: 0
Initial RMS Misfit: 17.86 Number of data ignored: 0
Initial RMS Misfit: 28.73 Number of data ignored: 0
Alpha: 428.484 RMS Misfit: 1.62 Roughness: 1.735
Alpha: 452.688 RMS Misfit: 4.68 Roughness: 6.958
Alpha: 430.648 RMS Misfit: 1.97 Roughness: 1.966
Alpha: 198.884 RMS Misfit: 1.24 Roughness: 2.403
Alpha: 199.889 RMS Misfit: 1.54 Roughness: 2.907
Alpha: 210.119 RMS Misfit: 4.18 Roughness: 10.993
Alpha: 92.314 RMS Misfit: 0.98 Roughness: 3.134
Step length set to 1.00000
Final RMS Misfit: 0.98
Alpha: 92.780 RMS Misfit: 1.23 Roughness: 4.065
Alpha: 97.529 RMS Misfit: 3.91 Roughness: 16.200
Cannot fit quadratic through step lengths
Alpha: 43.065 RMS Misfit: 1.02 Roughness: 5.479
Alpha: 45.269 RMS Misfit: 3.83 Roughness: 22.307
Final RMS Misfit: 0.98
Solution converged - Outputing results to file
Calculating sensitivity map
Processing dataset 2
End of data: Terminating
1/3 inversions completed Alpha: 19.989 RMS Misfit: 0.90 Roughness: 7.233
Alpha: 21.012 RMS Misfit: 3.85 Roughness: 28.839
Step length set to 1.00000
Final RMS Misfit: 0.90
Step length set to 1.00000
Final RMS Misfit: 3.83
Updated data weights
Iteration 2
All ok
All ok
Cannot fit quadratic through step lengths
Initial RMS Misfit: 2.66 Number of data ignored: 0
Final RMS Misfit: 0.90
Solution converged - Outputing results to file
Calculating sensitivity map
Processing dataset 2
End of data: Terminating
2/3 inversions completed Alpha: 27.097 RMS Misfit: 1.42 Roughness: 23.089
Alpha: 12.577 RMS Misfit: 1.01 Roughness: 31.706
Alpha: 5.838 RMS Misfit: 0.91 Roughness: 40.273
Step length set to 1.00000
Final RMS Misfit: 0.91
Final RMS Misfit: 1.01
Solution converged - Outputing results to file
Calculating sensitivity map
Processing dataset 2
End of data: Terminating
3/3 inversions completed
----------- END OF INVERSION IN // ----------
3/3 results parsed (3 ok; 0 failed)
All ok
00a4:err:rpc:I_RpcReceive we got fault packet with status 0x1c010003
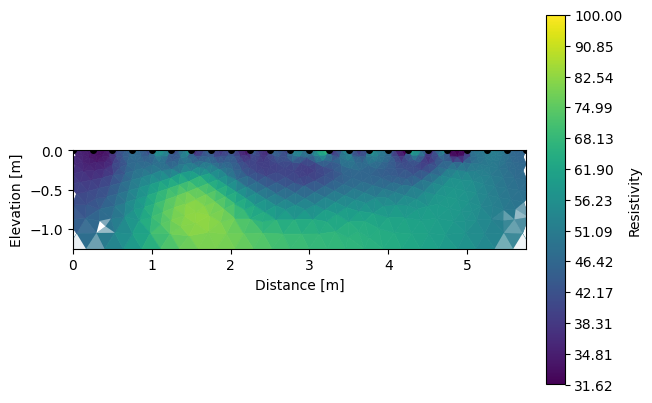
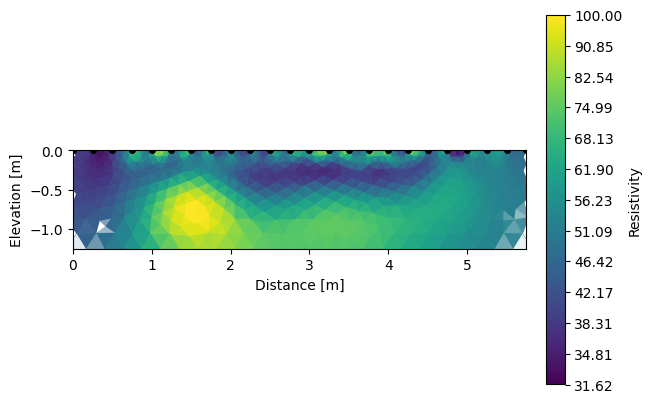
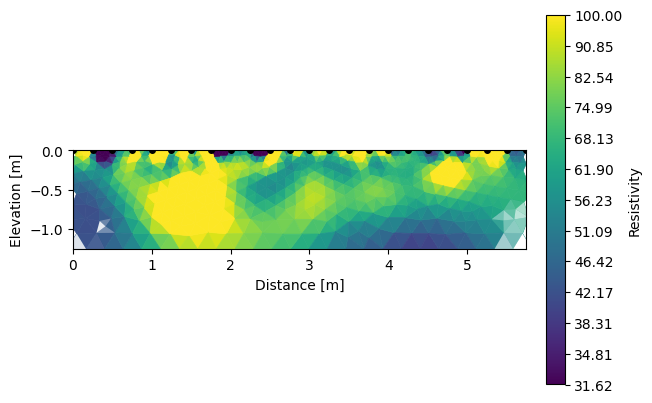
And each inverted section can be shown using the same function R2.showResults().
[5]:
k.showResults(index=0, vmin=1.5, vmax=2)
k.showResults(index=1, vmin=1.5, vmax=2)
k.showResults(index=2, vmin=1.5, vmax=2)
[ ]: