This page was generated from
jupyter-notebook/nb_3d-visualisation.ipynb.
Interactive online version:
 .
Download notebook.
.
Download notebook.
3D viewing
This notebook will show how to display 3D results with resipy in a notebook. Note that 3D viewing is based on the Python package: pyvista that needs to be installed (pip install pyvista).
[1]:
import sys
sys.path.append('../src')
from resipy import Project
import pyvista as pv
testdir = '../src/examples/'
API path = /media/jkl/data/phd/resipy/src/resipy
ResIPy version = 3.4.6
cR2.exe found and up to date.
R3t.exe found and up to date.
cR3t.exe found and up to date.
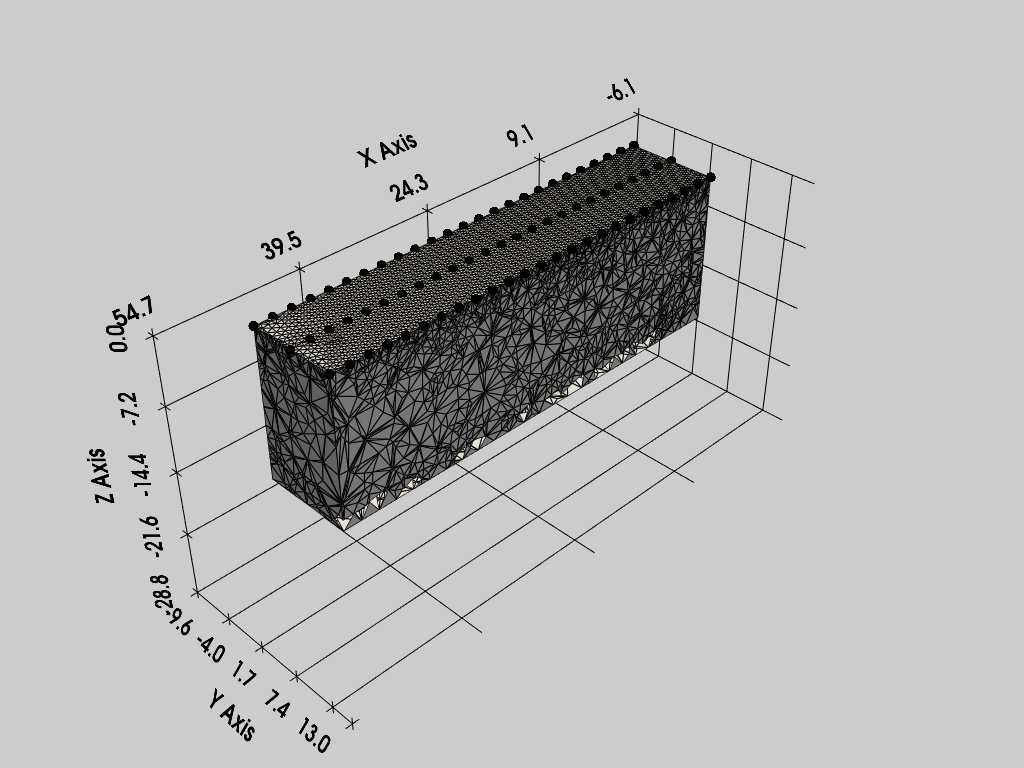
Create a Survey and Mesh, then display the mesh using a pyvista plotter object (which displays inline with the notebook).
[2]:
k = Project(typ='R3t')
k.createSurvey(testdir + 'dc-3d/protocol.dat', ftype='ProtocolDC')
k.importElec(testdir + 'dc-3d/elec.csv')
k.createMesh()
pl = pv.Plotter() # init pyvista plotter object
k.showMesh(ax=pl)
Working directory is: /media/jkl/data/phd/resipy/src/resipy
clearing dirname
0/644 reciprocal measurements found.
Creating tetrahedral mesh...done (62877 elements)
/media/jkl/data/phd/resipy/pyenv/lib/python3.10/site-packages/pyvista/jupyter/notebook.py:33: UserWarning: Failed to use notebook backend:
No module named 'trame'
Falling back to a static output.
warnings.warn(
Now invert the data
[3]:
k.invert()
Writing .in file and protocol.dat... done
--------------------- MAIN INVERSION ------------------
>> R 3 t E R T M o d e l v 2.31 <<
>> Date: 03-12-2023
>> My beautiful 3D survey
>> I n v e r s e S o l u t i o n S e l e c t e d <<
>> A d v a n c e d M e s h I n p u t <<
>> T e t r a h e d r a l E l e m e n t M e s h <<
>> Reading mesh file
>> Determining storage needed for finite element conductance matrix
>> Generating index array for finite element conductance matrix
>> Reading resistivity model from res0.dat
>> L o g - D a t a I n v e r s i o n <<
>> N o r m a l R e g u l a r i s a t i o n <<
>> Memory estimates:
For 1000 measurements the memory needed is: 0.521 Gb
For 2000 measurements the memory needed is: 1.024 Gb
For 5000 measurements the memory needed is: 2.534 Gb
For 10000 measurements the memory needed is: 5.049 Gb
>> Forming roughness matrix
>> Number of measurements read: 644
>> Total Memory required is: 0.342 Gb
Processing frame 1 - output to file f001.dat
Iteration 1
Initial RMS Misfit: 2.61 Number of data ignored: 0
Alpha: 21.523 RMS Misfit: 0.36 Roughness: 5.440
Step length set to 1.000
Final RMS Misfit: 0.36
Final RMS Misfit: 1.01
Solution converged - Outputing results to file
Calculating sensitivity map
End of data: Terminating
>> Program ended normally
1/1 results parsed (1 ok; 0 failed)
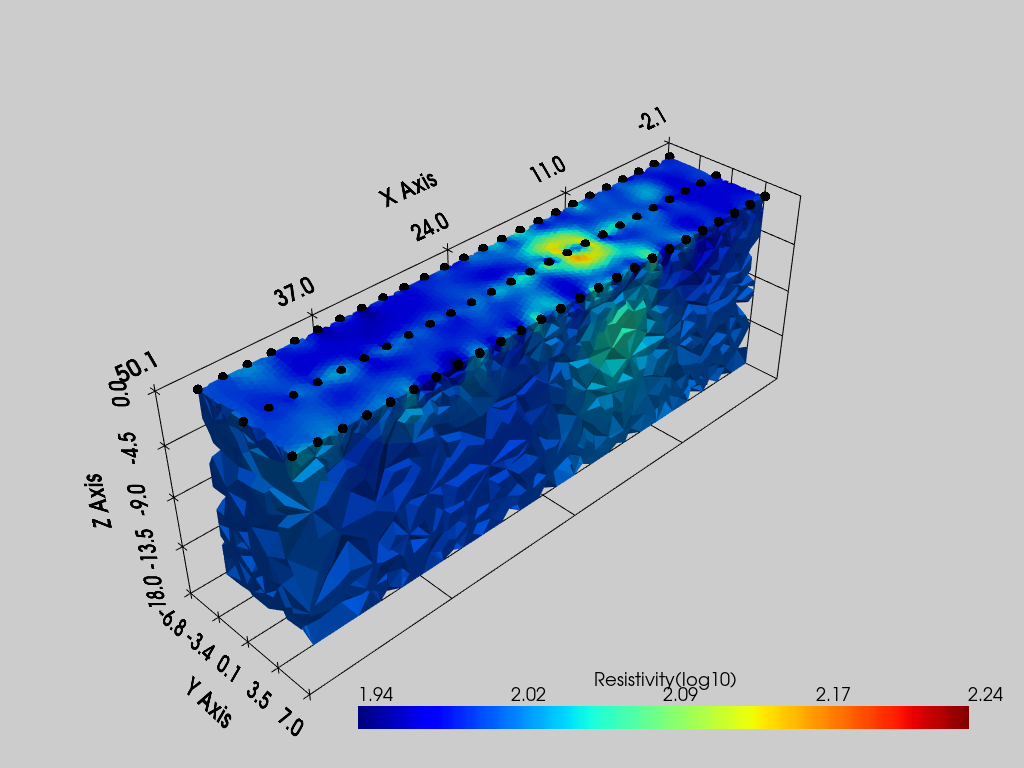
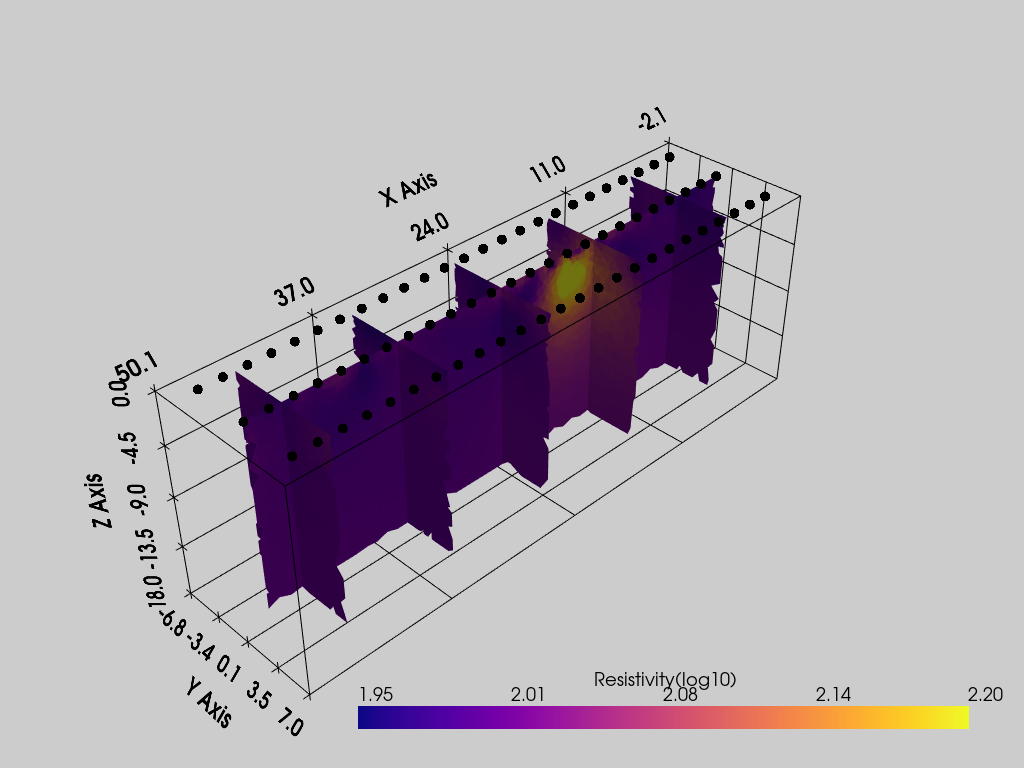
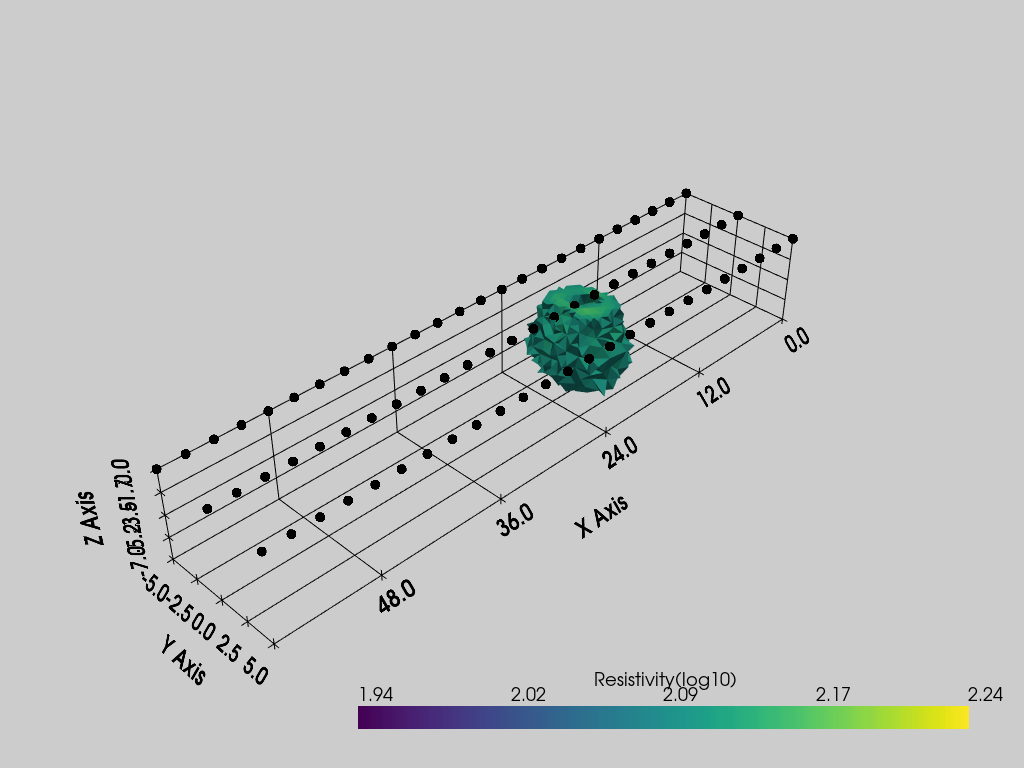
Options available
pvslices: a tuple of list, each list represent the coordinates of an slice orthogonal to X, Y and Z respectivelypvthreshold: a list of two values representing the minimum and maximum values between which to keep the cellspvcontour: a list of values at which draw isosurfacespvgrid: a bool, ifTrue, the grid is plotted
[4]:
pl = pv.Plotter()
k.showResults(ax=pl, color_map='jet')
/media/jkl/data/phd/resipy/pyenv/lib/python3.10/site-packages/pyvista/jupyter/notebook.py:33: UserWarning: Failed to use notebook backend:
No module named 'trame'
Falling back to a static output.
warnings.warn(
[5]:
pl = pv.Plotter()
k.showResults(ax=pl, pvslices=([5,15,25,35,45],[0],[]), pvgrid=True, vmin=1.95, vmax=2.2, color_map='plasma')
/media/jkl/data/phd/resipy/pyenv/lib/python3.10/site-packages/pyvista/jupyter/notebook.py:33: UserWarning: Failed to use notebook backend:
No module named 'trame'
Falling back to a static output.
warnings.warn(
[6]:
pl = pv.Plotter()
k.showResults(ax=pl, pvthreshold=[2.1, 2.2], pvgrid=True)
/media/jkl/data/phd/resipy/pyenv/lib/python3.10/site-packages/pyvista/jupyter/notebook.py:33: UserWarning: Failed to use notebook backend:
No module named 'trame'
Falling back to a static output.
warnings.warn(
[7]:
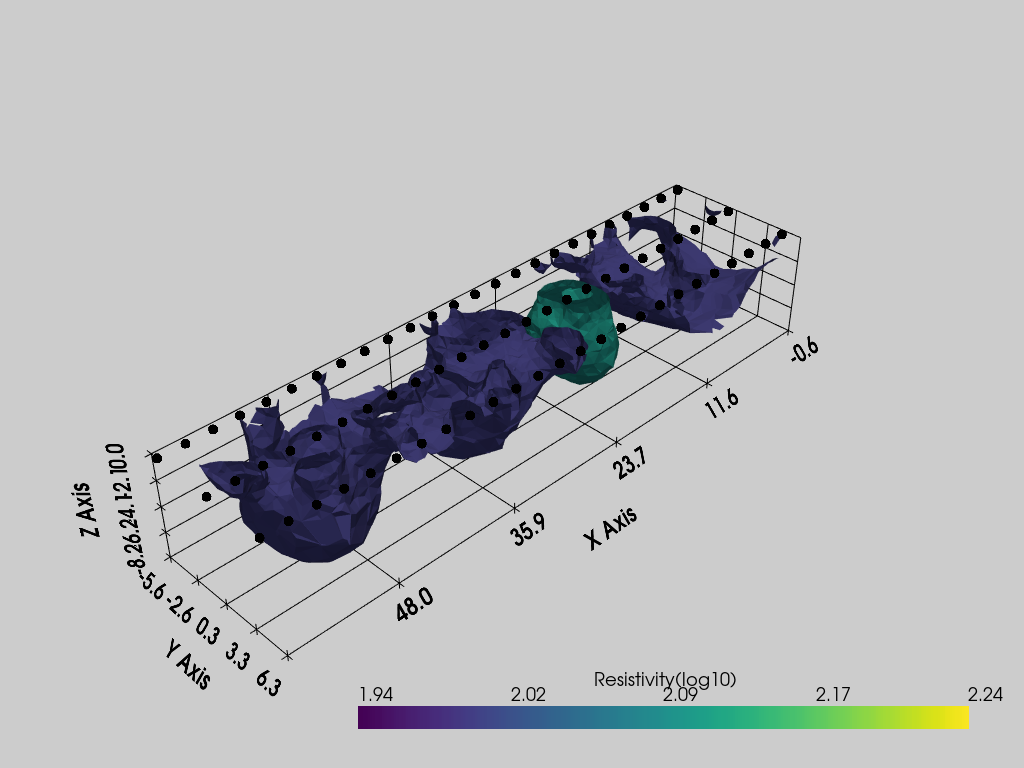
pl = pv.Plotter()
k.showResults(ax=pl, pvcontour=[2, 2.1], pvgrid=True)
/media/jkl/data/phd/resipy/pyenv/lib/python3.10/site-packages/pyvista/jupyter/notebook.py:33: UserWarning: Failed to use notebook backend:
No module named 'trame'
Falling back to a static output.
warnings.warn(
[ ]: