Forward modelling
In this tutorial, we will see how to use pyR2 API to do forward modelling.
[1]:
import warnings
warnings.filterwarnings('ignore')
import os
import sys
sys.path.append((os.path.relpath('../src'))) # add here the relative path of the API folder
import numpy as np # numpy for electrode generation
from resipy import Project
API path = /media/jkl/data/phd/resipy/src/resipy
ResIPy version = 3.4.6
cR2.exe found and up to date.
R3t.exe found and up to date.
cR3t.exe found and up to date.
[2]:
k = Project(typ='R2') # create R2 object
Working directory is: /media/jkl/data/phd/resipy/src/resipy
clearing dirname
First we need to design some electrodes. We will use numpy functions for this.
[3]:
elec = np.zeros((24,3))
elec[:,0] = np.arange(0, 24*0.5, 0.5) # with 0.5 m spacing and 24 electrodes
k.setElec(elec)
print(k.elec)
label x y z remote buried
0 1 0.0 0.0 0.0 False False
1 2 0.5 0.0 0.0 False False
2 3 1.0 0.0 0.0 False False
3 4 1.5 0.0 0.0 False False
4 5 2.0 0.0 0.0 False False
5 6 2.5 0.0 0.0 False False
6 7 3.0 0.0 0.0 False False
7 8 3.5 0.0 0.0 False False
8 9 4.0 0.0 0.0 False False
9 10 4.5 0.0 0.0 False False
10 11 5.0 0.0 0.0 False False
11 12 5.5 0.0 0.0 False False
12 13 6.0 0.0 0.0 False False
13 14 6.5 0.0 0.0 False False
14 15 7.0 0.0 0.0 False False
15 16 7.5 0.0 0.0 False False
16 17 8.0 0.0 0.0 False False
17 18 8.5 0.0 0.0 False False
18 19 9.0 0.0 0.0 False False
19 20 9.5 0.0 0.0 False False
20 21 10.0 0.0 0.0 False False
21 22 10.5 0.0 0.0 False False
22 23 11.0 0.0 0.0 False False
23 24 11.5 0.0 0.0 False False
Now let’s create a mesh.
[4]:
k.createMesh(typ='trian', show_output=False, res0=200) # let's create the mesh based on these electrodes position
k.showMesh()
Creating triangular mesh...done (1887 elements)
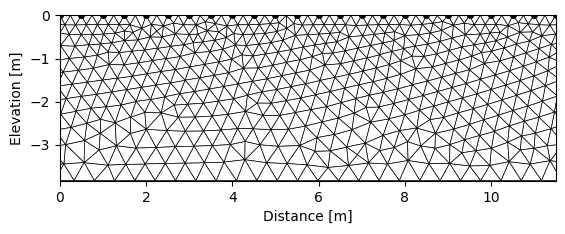
Based on this mesh, we can defined regions and assign them conductivities. There is an interactive way to do it when working outside of the jupyter notebook in interactive mode or GUI. Here we will see the pure API based way to do it using R2.addRegion().
[5]:
help(k.addRegion) # to display the help of the method
Help on method addRegion in module resipy.Project:
addRegion(xz, res0=100, phase0=1, blocky=False, fixed=False, ax=None, iplot=False) method of resipy.Project.Project instance
Add region according to a polyline defined by `xz` and assign it
the starting resistivity `res0`.
Parameters
----------
xz : array
Array with two columns for the x and y coordinates.
res0 : float, optional
Resistivity values of the defined area.
phase0 : float, optional
Read only if you choose the cR2 option. Phase value of the defined
area in mrad
blocky : bool, optional
If `True` the boundary of the region will be blocky if inversion
is block inversion.
fixed : bool, optional
If `True`, the inversion will keep the starting resistivity of this
region.
ax : matplotlib.axes.Axes
If not `None`, the region will be plotted against this axes.
iplot : bool, optional
If `True` , the updated mesh with the region will be plotted.
[6]:
k.addRegion(np.array([[2,-0.3],[2,-2],[3,-2],[3,-0.3],[2,-0.3]]), 50, iplot=True)
# first specify the path of the region and then its resistivity value in Ohm.m
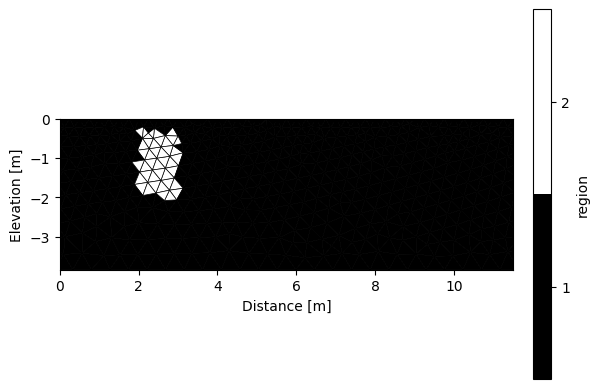
We then need to define the sequence that we will use. We can easily create a dipole-dipole sequence using R2.createSequence() or import one using R2.importSequence().
[7]:
help(k.createSequence) # don't hersitate to use the help to know more about each method
Help on method createSequence in module resipy.Project:
createSequence(params=[('dpdp1', 1, 8)], seqIdx=None, *kwargs) method of resipy.Project.Project instance
Creates a forward modelling sequence, see examples below for usage.
Parameters
----------
params : list of tuple, optional
Each tuple is the form (<array_name>, param1, param2, ...)
Types of sequences available are : 'dpdp1','dpdp2','wenner_alpha',
'wenner_beta', 'wenner_gamma', 'schlum1', 'schlum2', 'multigrad',
'custSeq'.
if 'custSeq' is chosen, param1 should be a string of file path to a .csv
file containing a custom sequence with 4 columns (a, b, m, n) containing
forward model sequence.
seqIdx: list of array like, optional
Each entry in list contains electrode indices (not label and string)
for a given electrode string which is to be sequenced. The advantage
of a list means that sequences can be of different lengths.
Examples
--------
>>> k = Project()
>>> k.setElec(np.c_[np.linspace(0,5.75, 24), np.zeros((24, 2))])
>>> k.createMesh(typ='trian')
>>> k.createSequence([('dpdp1', 1, 8), ('wenner_alpha', 1), ('wenner_alpha', 2)]) # dipole-dipole sequence
>>> k.createSequence([('custSeq', '<path to sequence file>/sequence.csv')]) # importing a custom sequence
>>> seqIdx = [[0,1,2,3],[4,5,6,7],[8,9,10,11,12]]
[8]:
k.createSequence([('dpdp1', 1, 10)]) # create a dipole-dipole of diple spacing of 1 (=skip 0) with 10 levels
print(k.sequence) # the sequence is stored inside the R2 object
165 quadrupoles generated.
[[ 1 2 3 4]
[ 2 3 4 5]
[ 3 4 5 6]
[ 4 5 6 7]
[ 5 6 7 8]
[ 6 7 8 9]
[ 7 8 9 10]
[ 8 9 10 11]
[ 9 10 11 12]
[10 11 12 13]
[11 12 13 14]
[12 13 14 15]
[13 14 15 16]
[14 15 16 17]
[15 16 17 18]
[16 17 18 19]
[17 18 19 20]
[18 19 20 21]
[19 20 21 22]
[20 21 22 23]
[21 22 23 24]
[ 1 2 4 5]
[ 2 3 5 6]
[ 3 4 6 7]
[ 4 5 7 8]
[ 5 6 8 9]
[ 6 7 9 10]
[ 7 8 10 11]
[ 8 9 11 12]
[ 9 10 12 13]
[10 11 13 14]
[11 12 14 15]
[12 13 15 16]
[13 14 16 17]
[14 15 17 18]
[15 16 18 19]
[16 17 19 20]
[17 18 20 21]
[18 19 21 22]
[19 20 22 23]
[20 21 23 24]
[ 1 2 5 6]
[ 2 3 6 7]
[ 3 4 7 8]
[ 4 5 8 9]
[ 5 6 9 10]
[ 6 7 10 11]
[ 7 8 11 12]
[ 8 9 12 13]
[ 9 10 13 14]
[10 11 14 15]
[11 12 15 16]
[12 13 16 17]
[13 14 17 18]
[14 15 18 19]
[15 16 19 20]
[16 17 20 21]
[17 18 21 22]
[18 19 22 23]
[19 20 23 24]
[ 1 2 6 7]
[ 2 3 7 8]
[ 3 4 8 9]
[ 4 5 9 10]
[ 5 6 10 11]
[ 6 7 11 12]
[ 7 8 12 13]
[ 8 9 13 14]
[ 9 10 14 15]
[10 11 15 16]
[11 12 16 17]
[12 13 17 18]
[13 14 18 19]
[14 15 19 20]
[15 16 20 21]
[16 17 21 22]
[17 18 22 23]
[18 19 23 24]
[ 1 2 7 8]
[ 2 3 8 9]
[ 3 4 9 10]
[ 4 5 10 11]
[ 5 6 11 12]
[ 6 7 12 13]
[ 7 8 13 14]
[ 8 9 14 15]
[ 9 10 15 16]
[10 11 16 17]
[11 12 17 18]
[12 13 18 19]
[13 14 19 20]
[14 15 20 21]
[15 16 21 22]
[16 17 22 23]
[17 18 23 24]
[ 1 2 8 9]
[ 2 3 9 10]
[ 3 4 10 11]
[ 4 5 11 12]
[ 5 6 12 13]
[ 6 7 13 14]
[ 7 8 14 15]
[ 8 9 15 16]
[ 9 10 16 17]
[10 11 17 18]
[11 12 18 19]
[12 13 19 20]
[13 14 20 21]
[14 15 21 22]
[15 16 22 23]
[16 17 23 24]
[ 1 2 9 10]
[ 2 3 10 11]
[ 3 4 11 12]
[ 4 5 12 13]
[ 5 6 13 14]
[ 6 7 14 15]
[ 7 8 15 16]
[ 8 9 16 17]
[ 9 10 17 18]
[10 11 18 19]
[11 12 19 20]
[12 13 20 21]
[13 14 21 22]
[14 15 22 23]
[15 16 23 24]
[ 1 2 10 11]
[ 2 3 11 12]
[ 3 4 12 13]
[ 4 5 13 14]
[ 5 6 14 15]
[ 6 7 15 16]
[ 7 8 16 17]
[ 8 9 17 18]
[ 9 10 18 19]
[10 11 19 20]
[11 12 20 21]
[12 13 21 22]
[13 14 22 23]
[14 15 23 24]
[ 1 2 11 12]
[ 2 3 12 13]
[ 3 4 13 14]
[ 4 5 14 15]
[ 5 6 15 16]
[ 6 7 16 17]
[ 7 8 17 18]
[ 8 9 18 19]
[ 9 10 19 20]
[10 11 20 21]
[11 12 21 22]
[12 13 22 23]
[13 14 23 24]
[ 1 2 12 13]
[ 2 3 13 14]
[ 3 4 14 15]
[ 4 5 15 16]
[ 5 6 16 17]
[ 6 7 17 18]
[ 7 8 18 19]
[ 8 9 19 20]
[ 9 10 20 21]
[10 11 21 22]
[11 12 22 23]
[12 13 23 24]]
Then comes the forward modelling part itself. The forward modelling will run R2, cR2, … in forward mode inside a fwd directory inside the working directory. The resulting apparent resistivity are then embeded inside a Survey object and directly available for inversion for instance.
[9]:
k.forward(noise=0.05, iplot=True) # forward modelling with 5 % noise added to the output
Writing .in file and mesh.dat... done
Writing protocol.dat... done
Running forward model...
>> R 2 R e s i s t i v i t y I n v e r s i o n v4.10 <<
>> D a t e : 03 - 12 - 2023
>> My beautiful survey
>> F o r w a r d S o l u t i o n S e l e c t e d <<
>> Determining storage needed for finite element conductance matrix
>> Generating index array for finite element conductance matrix
>> Reading start resistivity from resistivity.dat
Measurements read: 165 Measurements rejected: 0
>> Total Memory required is: 0.000 Gb
0/165 reciprocal measurements found.
All ok
/media/jkl/data/phd/resipy/pyenv/lib/python3.10/site-packages/numpy/lib/function_base.py:2458: DeprecationWarning: Conversion of an array with ndim > 0 to a scalar is deprecated, and will error in future. Ensure you extract a single element from your array before performing this operation. (Deprecated NumPy 1.25.)
res = asanyarray(outputs, dtype=otypes[0])
Forward modelling done.
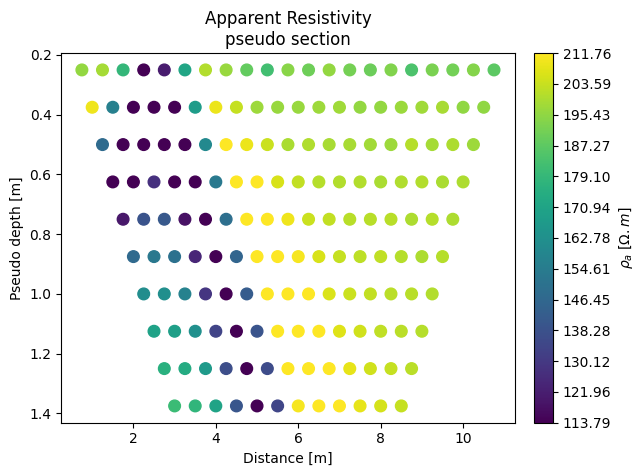
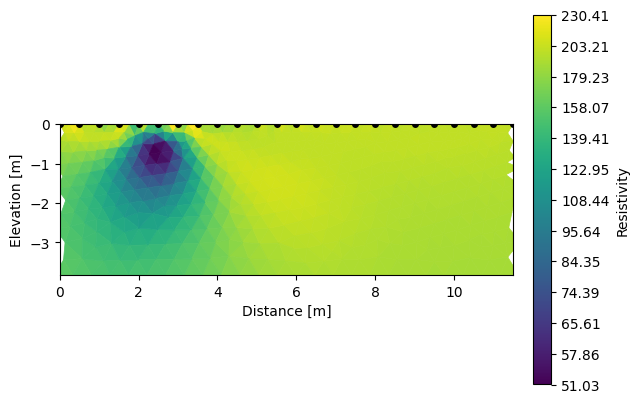
We can already see that the pseudo-section already show clearly the import on the region we defined. We can now invert these apparent resistivities. Inverting the forward models allow the user to see if the parameters of the surveys (the sequence and electrode spacing) were optimium to resolve the target. If needed he can change them and do the whole process again.
[10]:
k.invert()
Writing .in file and protocol.dat... All non fixed parameters reset to 100 Ohm.m and 0 mrad, as the survey to be inverted is from a forward model.
done
--------------------- MAIN INVERSION ------------------
>> R 2 R e s i s t i v i t y I n v e r s i o n v4.10 <<
>> D a t e : 03 - 12 - 2023
>> My beautiful survey
>> I n v e r s e S o l u t i o n S e l e c t e d <<
>> Determining storage needed for finite element conductance matrix
>> Generating index array for finite element conductance matrix
>> Reading start resistivity from res0.dat
>> R e g u l a r i s e d T y p e <<
>> L i n e a r F i l t e r <<
>> L o g - D a t a I n v e r s i o n <<
>> N o r m a l R e g u l a r i s a t i o n <<
>> D a t a w e i g h t s w i l l b e m o d i f i e d <<
Processing dataset 1
Measurements read: 165 Measurements rejected: 0
Geometric mean of apparent resistivities: 0.17448E+03
>> Total Memory required is: 0.003 Gb
Iteration 1
Initial RMS Misfit: 23.61 Number of data ignored: 0
Alpha: 1359.972 RMS Misfit: 3.12 Roughness: 0.995
Alpha: 631.243 RMS Misfit: 2.18 Roughness: 1.872
Alpha: 292.997 RMS Misfit: 1.48 Roughness: 2.993
Alpha: 135.997 RMS Misfit: 1.23 Roughness: 4.157
Alpha: 63.124 RMS Misfit: 1.31 Roughness: 5.331
Step length set to 1.00000
Final RMS Misfit: 1.23
Attempted to update data weights and caused overshoot
treating as converged
Solution converged - Outputing results to file
Calculating sensitivity map
Processing dataset 2
End of data: Terminating
1/1 results parsed (1 ok; 0 failed)
All ok
[11]:
k.showResults(index=0) # show the initial model
k.showResults(index=1, sens=False) # show the inverted model
ERROR: No sensitivity attribute found
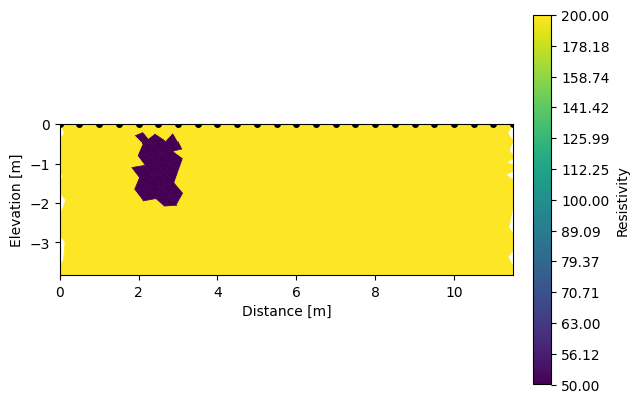
In a nutshell
[12]:
k = Project(typ='R2')
# defining electrode array
x = np.zeros((24, 3))
x[:,0] = np.arange(0, 24*0.5, 0.5)
k.setElec(elec)
# creating mesh
k.createMesh()
# add region
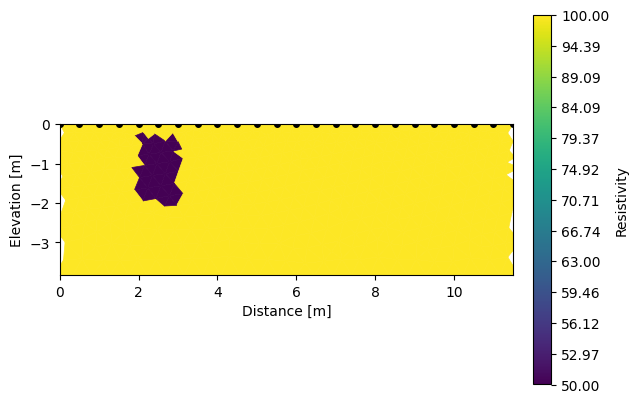
k.addRegion(np.array([[2,-0.3],[2,-2],[3,-2],[3,-0.3],[2,-0.3]]), 50)
# define sequence
k.createSequence([('dpdp1', 1, 10)])
# forward modelling
k.forward(noise=0.0)
# inverse modelling based on forward results
k.invert()
# show the initial and recovered section
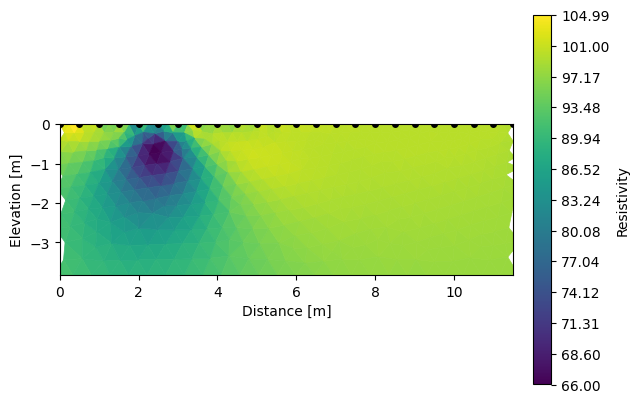
k.showResults(index=0) # initial
k.showResults(index=1, sens=False) # recovered
Working directory is: /media/jkl/data/phd/resipy/src/resipy
clearing dirname
Creating triangular mesh...done (1887 elements)
165 quadrupoles generated.
Writing .in file and mesh.dat... done
Writing protocol.dat... done
Running forward model...
>> R 2 R e s i s t i v i t y I n v e r s i o n v4.10 <<
>> D a t e : 03 - 12 - 2023
>> My beautiful survey
>> F o r w a r d S o l u t i o n S e l e c t e d <<
>> Determining storage needed for finite element conductance matrix
>> Generating index array for finite element conductance matrix
>> Reading start resistivity from resistivity.dat
Measurements read: 165 Measurements rejected: 0
>> Total Memory required is: 0.000 Gb
0/165 reciprocal measurements found.
All ok
/media/jkl/data/phd/resipy/pyenv/lib/python3.10/site-packages/numpy/lib/function_base.py:2458: DeprecationWarning: Conversion of an array with ndim > 0 to a scalar is deprecated, and will error in future. Ensure you extract a single element from your array before performing this operation. (Deprecated NumPy 1.25.)
res = asanyarray(outputs, dtype=otypes[0])
Forward modelling done.Writing .in file and protocol.dat... All non fixed parameters reset to 100 Ohm.m and 0 mrad, as the survey to be inverted is from a forward model.
done
--------------------- MAIN INVERSION ------------------
>> R 2 R e s i s t i v i t y I n v e r s i o n v4.10 <<
>> D a t e : 03 - 12 - 2023
>> My beautiful survey
>> I n v e r s e S o l u t i o n S e l e c t e d <<
>> Determining storage needed for finite element conductance matrix
>> Generating index array for finite element conductance matrix
>> Reading start resistivity from res0.dat
>> R e g u l a r i s e d T y p e <<
>> L i n e a r F i l t e r <<
>> L o g - D a t a I n v e r s i o n <<
>> N o r m a l R e g u l a r i s a t i o n <<
>> D a t a w e i g h t s w i l l b e m o d i f i e d <<
Processing dataset 1
Measurements read: 165 Measurements rejected: 0
Geometric mean of apparent resistivities: 0.93704E+02
>> Total Memory required is: 0.003 Gb
Iteration 1
Initial RMS Misfit: 3.71 Number of data ignored: 0
Alpha: 1109.630 RMS Misfit: 1.29 Roughness: 0.203
Alpha: 515.045 RMS Misfit: 0.91 Roughness: 0.379
Step length set to 1.00000
Final RMS Misfit: 0.91
Final RMS Misfit: 1.01
Solution converged - Outputing results to file
Calculating sensitivity map
Processing dataset 2
End of data: Terminating
1/1 results parsed (1 ok; 0 failed)
ERROR: No sensitivity attribute found
All ok
[ ]: