Global sensitivity analysis
Global sensitivity analysis is a Monte Carlo based method to rank the importance of parameters in a given modelling problem. As opposed to local senstivity analysis, it does not require the construction of the Jacobian, making it a flexible tool to evaluate complex problems.
Global sensitivty analysis is available in mainly uncertainty quantificaiton packages, as well as some flow and transport programs (e.g. iTOUGH2). GSA is also very popular in catchment modelling and civil engineering/risk analysis problems.
Some GSA work in hydrogeophysics (mainly by Berkeley Lab):
coupled hydrological-thermal-geophysical inversion (Tran et al 2017) > Nicely show how to simplify (i.e. reduce the number of parameters) for a very complex, highly coupled problem
making sense of global senstivity analysis (Wainwright et al 2014) > Very good review article
Sensitivity analysis of environmental models (Pianosi et al 2014) > A systematic review, includes GLUE and RSA
hydrogeology of a nuclear site in the Paris Basin (Deman et al 2016) > A different GSA method was used instead here to look at the low probability breakthrough events
Global Sensitivity and Data-Worth Analyses in iTOUGH2 User’s Guide (Wainwright et al 2016) > An useful manual if you want to learn about the details of setting up a probllem.
In this tutorial, we will see how to link the ResIPy API and SALib for senstivity analysis. Two key elements of SA are (i) forward modelling (Monte Carlo runs) and (ii) specifying the parameter ranges. This notebook will showcase of the use of the Method of Morris, which is known for its relatively small computational cost. This tutorial is modified from the one posted on https://github.com/SALib/SATut to demonstrate its coupling with ResIPy
Morris sensitivity method
The Morris one-at-a-time (OAT) method (Morris, 1991) can be considered as an extension of the local sensitivity method. Each parameter range is scaled to the unit interval [0, 1] and partitioned into \((p−1)\) equally-sized intervals. The reference value of each parameter is selected randomly from the set \({0, 1/(p−1), 2/(p−1), …, 1−Δ}\). The fixed increment \(Δ=p/{2(p−1)}\) is added to each parameter in random order to compute the elementary effect (\(EE\)) of \(x_i\)
We compute three statistics: the mean \(EE\), standard deviation (STD) of \(EE\), and mean of absolute \(EE\). * mean EE (:math:`mu`) represents the average effect of each parameter over the parameter space, the mean EE can be regarded as a global sensitivity measure. * mean |EE| (:math:`mu*`) is used to identify the non-influential factors, * STD of EE (:math:`sigma`) is used to identify nonlinear and/or interaction effects. (The standard error of mean (SEM) of EE, defined as \(SEM=STD/r^{0.5}\), is used to calculate the confidence interval of mean EE (Morris, 1991))
This cell is copied from (Wainwright et al 2014)
Importing libraries
[13]:
import warnings
warnings.filterwarnings('ignore')
import os
import sys
sys.path.append('../src') # add here the relative path of the API folder
from resipy import Project
import numpy as np # numpy for electrode generation
import pandas as pd
from IPython.utils import io # suppress R2 outputs during MC runs
The SALib package
SALib is a free open-source Python library
If you use Python, you can install it by running the command
pip install SALib
Documentation is available online and you can also view the code on Github.
The library includes: * Sobol Sensitivity Analysis (Sobol 2001, Saltelli 2002, Saltelli et al. 2010) * Method of Morris, including groups and optimal trajectories (Morris 1991, Campolongo et al. 2007) * Fourier Amplitude Sensitivity Test (FAST) (Cukier et al. 1973, Saltelli et al. 1999) * Delta Moment-Independent Measure (Borgonovo 2007, Plischke et al. 2013) * Derivative-based Global Sensitivity Measure (DGSM) (Sobol and Kucherenko 2009) * Fractional Factorial Sensitivity Analysis (Saltelli et al. 2008)
[2]:
# import the packages
from SALib.sample import morris as ms
from SALib.analyze import morris as ma
from SALib.plotting import morris as mp
Create ERT forward problem with ResIPy
In the code below, created a Project forward problem to be analyzed
[3]:
k = Project(typ='R2')
elec = np.zeros((24,3))
elec[:,0] = np.arange(0, 24*0.5, 0.5) # with 0.5 m spacing and 24 electrodes
k.setElec(elec)
#print(k.elec)
# defining electrode array
x = np.zeros((24, 3))
x[:,0] = np.arange(0, 24*0.5, 0.5)
k.setElec(elec)
# creating mesh
k.createMesh(res0=20)
# add region
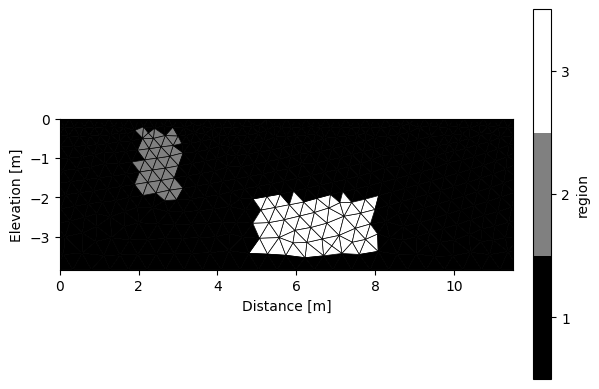
k.addRegion(np.array([[2,-0.3],[2,-2],[3,-2],[3,-0.3],[2,-0.3]]), 50)
k.addRegion(np.array([[5,-2],[5,-3.5],[8,-3.5],[8,-2],[5,-2]]), 500)
# define sequence
k.createSequence([('dpdp1', 1, 10)])
# forward modelling
k.forward(noise=0.025)
# read results
fwd_dir = os.path.relpath('../src/resipy/invdir/fwd')
obs_data = np.loadtxt(os.path.join(fwd_dir, 'R2_forward.dat'),skiprows =1)
obs_data = obs_data[:,6]
# plot
k.showMesh()
Working directory is: /media/jkl/data/phd/resipy/src/resipy
clearing dirname
Creating triangular mesh...done (1887 elements)
165 quadrupoles generated.
Writing .in file and mesh.dat... done
Writing protocol.dat... done
Running forward model...
>> R 2 R e s i s t i v i t y I n v e r s i o n v4.10 <<
>> D a t e : 03 - 12 - 2023
>> My beautiful survey
>> F o r w a r d S o l u t i o n S e l e c t e d <<
>> Determining storage needed for finite element conductance matrix
>> Generating index array for finite element conductance matrix
>> Reading start resistivity from resistivity.dat
Measurements read: 165 Measurements rejected: 0
>> Total Memory required is: 0.000 Gb
0/165 reciprocal measurements found.
Forward modelling done.
All ok
/media/jkl/data/phd/resipy/pyenv/lib/python3.10/site-packages/numpy/lib/function_base.py:2458: DeprecationWarning: Conversion of an array with ndim > 0 to a scalar is deprecated, and will error in future. Ensure you extract a single element from your array before performing this operation. (Deprecated NumPy 1.25.)
res = asanyarray(outputs, dtype=otypes[0])
Option to view resistivity fields with pyvista
[4]:
import pyvista as pv
mesh = pv.read(os.path.join(fwd_dir, 'forward_model.vtk'))
mesh.plot(scalars='Resistivity(log10)',notebook=True)
mesh
[4]:
| Header | Data Arrays | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
[5]:
import pyvista as pv
plotter = pv.Plotter()
mesh = pv.read(os.path.join(fwd_dir, 'forward_model.vtk'))
plotter.add_mesh(mesh,cmap="viridis" ,stitle="Resistivity(log10)") #scalars='Resistivity(log10)'
#plotter.show_axes()
_ = plotter.show_bounds(grid='front', location='outer', all_edges=True, xlabel="X [m]",ylabel="Y [m]")
plotter.add_text('pyvista example')
#plotter.update_scalar_bar_range([-2000,2000], name="Resistivity(log10)")
#plotter.add_mesh(mesh, cmap="bone", opacity="linear", stitle="Linear Opacity")
plotter.view_xy()
plotter.show()
#plotter.show(screenshot='test.png')
Define a problem file
In the code below, a problem file is used to define the parameters and their ranges we wish to explore, which corresponds to the following table:
Parameter |
Range |
Description |
|---|---|---|
rho0 [ohm m] |
10 [1] |
background |
rho1 [ohm m] |
10 [2] |
inclusion A |
rho2 [ohm m] |
10 [3] |
inclusion B |
[6]:
morris_problem = {
'num_vars': 3,
# These are their names
'names': ['rho1', 'rho2', 'rho3'], # can add z1 z2 etc.
# Plausible ranges over which we'll move the variables
'bounds': [[0.5,3.5], # log10 of rho (ohm m)
[0.5,3.5],
[0.5,3.5]#,
# [-3,-1],
# [-7,-4],
],
# I don't want to group any of these variables together
'groups': None
}
Generate a Sample
We then generate a sample using the morris.sample() procedure from the SALib package.
[7]:
number_of_trajectories = 20
sample = ms.sample(morris_problem, number_of_trajectories, num_levels=10)
len(sample)
print(sample[79,:])
[1.16666667 1.5 1.83333333]
Run the sample through the monte carlo procedure in R2
Great! You have defined your problem and have created a series of input files for forward runs. Now you need to run R2 for each of them to obtain their ERT responses.
For this example, each sample takes a few seconds to run on a PC.
[8]:
#%%capture
simu_ensemble = np.zeros((len(obs_data),len(sample)))
for ii in range(0, len(sample)):
with io.capture_output() as captured: # suppress inline output from ResIPy
# creating mesh
k.createMesh(res0=10**sample[ii,0]) # need to use more effective method, no need to create mesh every time
# add region
k.addRegion(np.array([[2,-0.3],[2,-2],[3,-2],[3,-0.3],[2,-0.3]]), 10**sample[ii,1])
k.addRegion(np.array([[5,-2],[5,-3.5],[8,-3.5],[8,-2],[5,-2]]), 10**sample[ii,2])
# forward modelling
k.forward(noise=0.025, iplot = False)
out_data = np.loadtxt(os.path.join(fwd_dir, 'R2_forward.dat'),skiprows =1)
simu_ensemble[:,ii] = out_data[:,6]
print("Running sample",ii+1)
All ok
Running sample 1
All ok
Running sample 2
All ok
Running sample 3
All ok
Running sample 4
All ok
Running sample 5
All ok
Running sample 6
All ok
Running sample 7
All ok
Running sample 8
All ok
Running sample 9
All ok
Running sample 10
All ok
Running sample 11
All ok
Running sample 12
All ok
Running sample 13
All ok
Running sample 14
All ok
Running sample 15
All ok
Running sample 16
All ok
Running sample 17
All ok
Running sample 18
All ok
Running sample 19
All ok
Running sample 20
All ok
Running sample 21
All ok
Running sample 22
All ok
Running sample 23
All ok
Running sample 24
All ok
Running sample 25
All ok
Running sample 26
All ok
Running sample 27
All ok
Running sample 28
All ok
Running sample 29
All ok
Running sample 30
All ok
Running sample 31
All ok
Running sample 32
All ok
Running sample 33
All ok
Running sample 34
All ok
Running sample 35
All ok
Running sample 36
All ok
Running sample 37
All ok
Running sample 38
All ok
Running sample 39
All ok
Running sample 40
All ok
Running sample 41
All ok
Running sample 42
All ok
Running sample 43
All ok
Running sample 44
All ok
Running sample 45
All ok
Running sample 46
All ok
Running sample 47
All ok
Running sample 48
All ok
Running sample 49
All ok
Running sample 50
All ok
Running sample 51
All ok
Running sample 52
All ok
Running sample 53
All ok
Running sample 54
All ok
Running sample 55
All ok
Running sample 56
All ok
Running sample 57
All ok
Running sample 58
All ok
Running sample 59
All ok
Running sample 60
All ok
Running sample 61
All ok
Running sample 62
All ok
Running sample 63
All ok
Running sample 64
All ok
Running sample 65
All ok
Running sample 66
All ok
Running sample 67
All ok
Running sample 68
All ok
Running sample 69
All ok
Running sample 70
All ok
Running sample 71
All ok
Running sample 72
All ok
Running sample 73
All ok
Running sample 74
All ok
Running sample 75
All ok
Running sample 76
All ok
Running sample 77
All ok
Running sample 78
All ok
Running sample 79
Running sample 80
All ok
Factor Prioritisation
We’ll run a sensitivity analysis of the power module to see which is the most influential parameter.
The results parameters are called mu, sigma and mu_star.
Mu is the mean effect caused by the input parameter being moved over its range.
Sigma is the standard deviation of the mean effect.
Mu_star is the mean absolute effect.
The higher the mean absolute effect for a parameter, the more sensitive/important it is*
[9]:
# Define an objective function: here I use the error weighted rmse
def obj_fun(sim,obs,noise):
y = np.divide(sim-obs,noise) # weighted data misfit
y = np.sqrt(np.inner(y,y))
return y
output = np.zeros((1,len(sample)))
for ii in range(0, len(sample)):
output[0,ii] = obj_fun(simu_ensemble[:,ii],obs_data,0.025*obs_data) # assume 2.5% noise in the data
# Store the results for plotting of the analysis
Si = ma.analyze(morris_problem, sample, output, print_to_console=False)
print("{:20s} {:>7s} {:>7s} {:>7s}".format("Name", "mean(EE)", "mean(|EE|)", "std(EE)"))
for name, s1, st, mean in zip(morris_problem['names'],
Si['mu'],
Si['mu_star'],
Si['sigma']):
print("{:20s} {:=7.3f} {:=7.3f} {:=7.3f}".format(name, s1, st, mean))
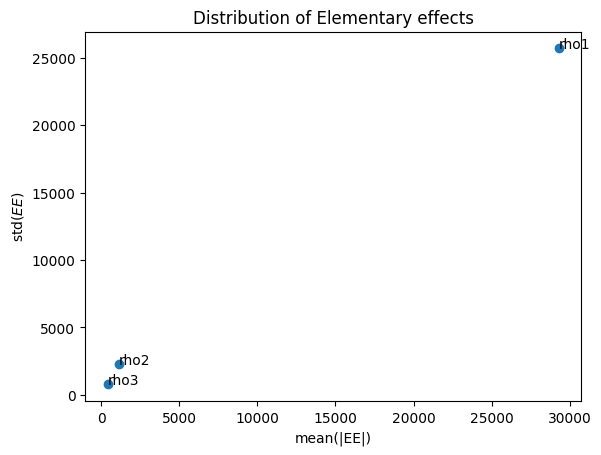
Name mean(EE) mean(|EE|) std(EE)
rho1 29271.700 29271.700 25700.482
rho2 1133.239 1146.483 2255.792
rho3 413.524 428.156 766.059
[10]:
# make a plot
import matplotlib.pyplot as plt
import numpy as np
fig, ax = plt.subplots()
ax.scatter(Si['mu_star'],Si['sigma'])
#ax.plot(Si['mu_star'],2*Si['sigma']/np.sqrt(number_of_trajectories),'--',alpha=0.5)
#ax.plot(np.array([0,Si['mu_star'][0]]),2*np.array([0,Si['sigma'][0]/np.sqrt(number_of_trajectories)]),'--',alpha=0.5)
plt.title('Distribution of Elementary effects')
plt.xlabel('mean(|EE|)')
plt.ylabel('std($EE$)')
for i, txt in enumerate(Si['names']):
ax.annotate(txt, (Si['mu_star'][i], Si['sigma'][i]))
# higher mean |EE|, more important factor
# line within the dashed envelope means nonlinear or interaction effects dominant
Summary
Sensitivity analysis helps you:
Think through your assumptions
Quantify uncertainty
Focus on the most influential uncertainties first
Learn Python
Similar packages to SALib for other languages/programmes:
‘sensitivity’ package for R (Michael Tso used it for GSA in his leak detection paper)
[11]:
# run this so that a navigation sidebar will bee generated when exporting this notebook as HTML
#%%javascript
#$('<div id="toc"></div>').css({position: 'fixed', top: '120px', left: 0}).appendTo(document.body);
#$.getScript('https://kmahelona.github.io/ipython_notebook_goodies/ipython_notebook_toc.js');
[12]:
help(k.addRegion)
Help on method addRegion in module resipy.Project:
addRegion(xz, res0=100, phase0=1, blocky=False, fixed=False, ax=None, iplot=False) method of resipy.Project.Project instance
Add region according to a polyline defined by `xz` and assign it
the starting resistivity `res0`.
Parameters
----------
xz : array
Array with two columns for the x and y coordinates.
res0 : float, optional
Resistivity values of the defined area.
phase0 : float, optional
Read only if you choose the cR2 option. Phase value of the defined
area in mrad
blocky : bool, optional
If `True` the boundary of the region will be blocky if inversion
is block inversion.
fixed : bool, optional
If `True`, the inversion will keep the starting resistivity of this
region.
ax : matplotlib.axes.Axes
If not `None`, the region will be plotted against this axes.
iplot : bool, optional
If `True` , the updated mesh with the region will be plotted.